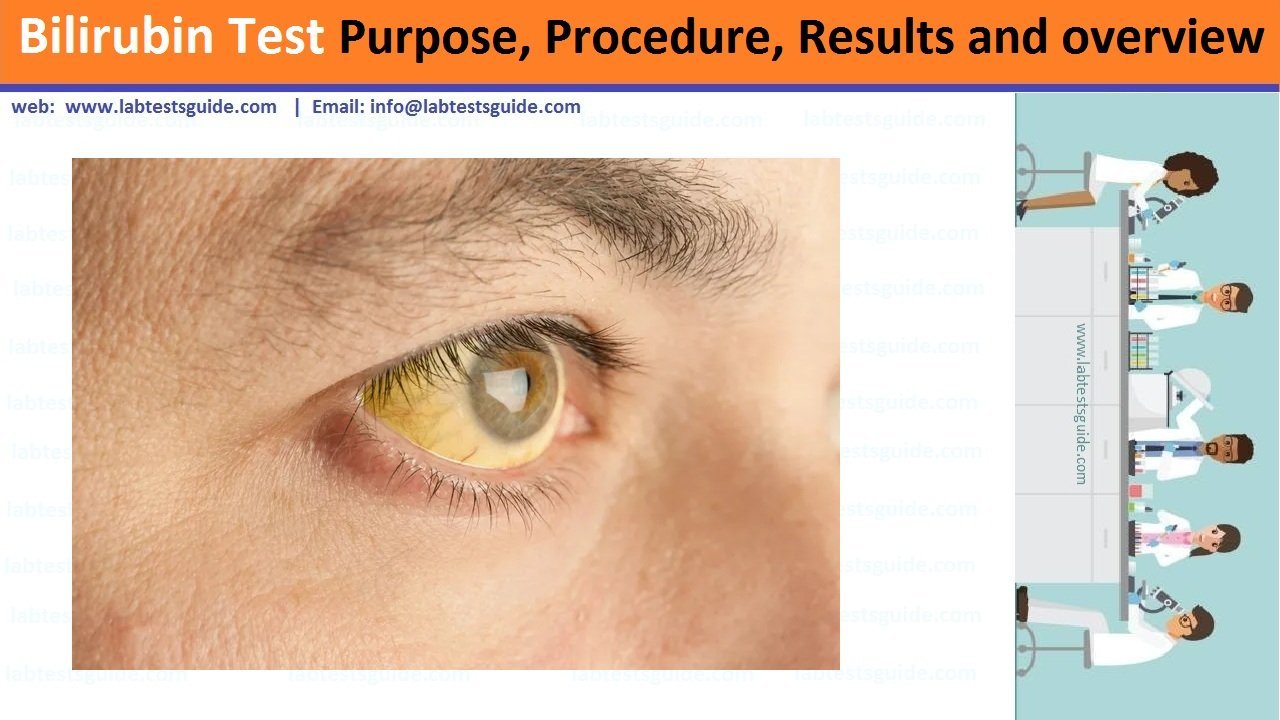
PCR amplification is a popular method used to amplify the short DNA fragments, and also called “Molecular photocopying”. PCR is an acronym used for Polymerase chain reaction. Kerry Mullis was the first scientist, who introduced PCR with its remarkable applicability in genetic and molecular biology.

It is a modern, inexpensive, and rapid (requires few hours) techniques of amplifying specific DNA sequences, while the traditional method was quite a time taking (requires several days or a week). Polymerase chain reaction can yield million to billion copies of the desired DNA sequence.
Content: PCR Amplification
Definition of PCR
PCR or polymerase chain reaction can define as in vitro technique of preparing around million copies of target DNA sequence through consecutive stages of heat denaturation, primer annealing, and primer extension. It is an advanced molecular technology that revolutionized the study of DNA more in medical and biological research like DNA mapping, DNA fingerprinting, forensics, detection of microorganisms, etc.
PCR aims to yield more copies of desired DNA sequence or provides different scopes in studying the DNA. For instance, DNA amplified by PCR can be used in different ways like DNA sequencing, DNA cloning, etc.
PCR Set-up
The polymerase chain reaction is a molecular method, which set-up requires:
- DNA template
- Primers (short stretches of DNA)
- DNA precursors
- Magnesium ion
- Taq polymerase enzyme
- Buffer solution
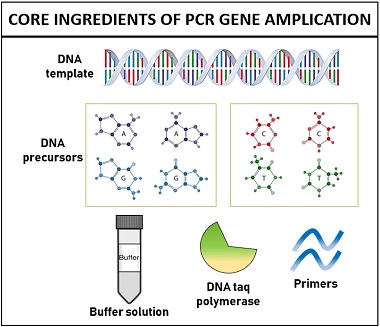
DNA template
It is the most important component of PCR, whose concentration and quality determines the success of amplification. The concentration of DNA template should be within 30 ng-50 ng, i.e. not too little or too high. Other than this, DNA template should be free of chemical contaminants that can inhibit the gene amplification.
DNA precursors
These are also known as deoxynucleoside triphosphates (dNTPs) including dATP, dCTP, dGTP, and dTTP to construct the new DNA strands. DNA precursors function as the building blocks of DNA, so they are added equivalently (generally 0.2 mM) for optimal base incorporation.
Magnesium ion
Mg2+ acts as a cofactor of DNA polymerase that facilitates the incorporation of DNA precursors during polymerization. The magnesium ions associates with an enzyme Taq polymerase to bring about the formation of phosphodiester bond between the primer terminus and the phosphate group of dNTPs. It is utilized through MgCl2 solution added to the PCR mixture, which concentration is optimized within a range of 1–4 mM. Its high and low concentration can affect the productivity of PCR.
Taq polymerase enzyme
It is a DNA polymerase enzyme obtained from Thermus aquaticus (Heat-loving bacteria) that thrives in hot springs and hydrothermal vents. Taq DNA polymerase possesses high thermostability (remains stable above 80°C).
It is most active at a temperature of 70°C, at which it can add on approximately 60 bases per second. The heat-stability of Taq polymerase is ideal for PCR, as the template DNA is subjected to thermal cycling to get new DNA strands.
PCR primers
These are the short pieces of single-stranded DNA (15-30 bp long) that synthesizing the new DNA strand. PCR makes the use of two primers. For its design, some information prior to the desired DNA sequence from the target DNA is required to design two primers.
The PCR primers are selectively designed relative to the desired DNA sequence in a template DNA. Its just attach oppositely towards the edges of the intended DNA sequence via complementary base pairing. Once, it binds to the desired DNA sequence, then the sequence can be lengthened by the DNA polymerase, and the target DNA sequence gets copied. Ideal properties of primers include:
- Primer length: Its length should not be too short or long, as it might hybridize the non-target sites or target DNA, respectively.
- Base composition: It should be even and the GC content must lie in between 40-60%.
- Nature of sequences: Any self-complementary sequences must be avoided to minimize mispriming or non-selective priming.
- Melting temperature: Tm value for the two primers should be within 5°C.
PCR buffers
It maintains the pH alternations in the sample to the optimal pH (8.0 and 9.5) required for the enzymatic action of Taq DNA polymerase, which is optimized by adding Tris-HCl.
- KCl buffer: Here, the potassium or K+ ion promotes primer annealing.
- (NH4)2SO4 buffer: It is used as an alternative to KCl buffer. Here, the ammonium or NH4+ ion enhance specificity by its destabilizing property of weak H-bonds formed between the mismatched primer and template DNA during DNA elongation.
For optimal PCR yields and robust enzyme activity of Taq DNA polymerases, one should use the buffer provided with the DNA polymerase.
PCR Amplification Cycle
The PCR reactants mentioned above are assembled in a tube and subjected to repetitive thermocycling to yield new DNA strands. PCR cycle involves three sequential stages:
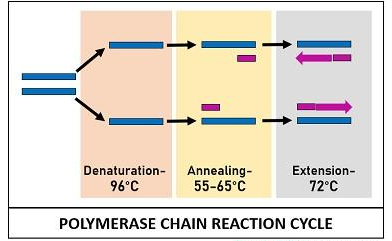
Denaturation
It is an initial stage where the solution is subjected to a high temperature of 96°C. This reaction cause separation or denaturation of target ds-DNA into template ss-DNA. Denaturation of the target DNA occurs via disintegration of H-bonds between the bases.
The temperature is maintained for a certain time to establish complete separation of the DNA, so that the single-stranded or separated DNA can be used as a template for the synthesis of new DNA.
Annealing
It is a second stage when the sample is cooled up to 55-65°C of temperature. This reaction facilitates the binding of the two primers to their complementary sequences of the template DNA via H-bond.
Primers are the short DNA sequences that act as a starting point to polymerize or extend the DNA. During this step, the two primers attach towards the edge of the template DNA, opposite to each other.
Extension
This is the last stage when the sample is again subjected to high temperature (72°C). At this temperature, an enzyme DNA taq-polymerase streches out the short sequenced primers by incorporating DNA bases complementary to the template strand. Taq polymerase adds DNA precursors or nucleotide bases from the 5’ to 3’ direction. This step results in the formation of a new DNA strands, which synthesis is determined by the length of DNA sequence that has to be amplified.
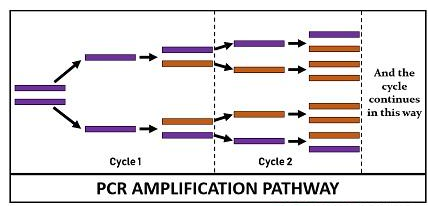
Finally, the reaction chamber is cooled up to 4–15 °C for short-term storage of the PCR products. The reaction cycle of PCR repeats 25-35 times to produce millions to the billions of copies. Generally, the reaction cycle takes 22-44 hours to complete but depends on the DNA length, the concentration of PCR ingredients, the working efficiency of the instrument, and more other factors.
Advantages
- It is a simple molecular technique that can be handled easily.
- PCR amplification gives a large number of PCR products rapidly.
- It is a selective amplification technique, in which the forward and reverse primers selectively binds to the complementary sequence of the template DNA.
- The process is carried out in a closed system, which minimizes the chances of contamination.
- It becomes an important tool to get better yield, which in turn can be used in many research purposes.
Limitations
- Information about the target DNA is required to design primers for selective amplification.
- Optimization is quite difficult in the PCR set-up that can give misleading results, so it requires a sterile room for reagent preparation.
- DNA polymerase prone to error (about 40%), and may yield mutated DNA after 20 reaction cycles. Also, multiple primers can sometimes cause cross-hybridization and can give misleading results.
Conclusion
Therefore, we can conclude that PCR gene amplification aims to double the PCR product in each cycle that is achieved by 100% reaction efficiency. PCR produces exponential copies of DNA throughout the reaction cycle. A single copy of DNA can yield up to one billion copies via 30 rounds of amplification. Its working efficiency depends upon the optimization of the reagents used in the PCR set up.
Related Articles:
RELATED POSTS
View all